School Seminar: Dr Clare Mahon; Durham University
Wednesday, 6 April 4:00pm – 5:00pm
This seminar will be delivered in Online Zoom Please email chemistry.researchsupport@sydney.edu.au for zoom link and password.
Speaker: Dr Clare Mahon; Durham University
Host: Prof. Liz New
Title: Glycopolymer sensing arrays to detect pathoadaptations in Pseudomonas aeruginosa
Abstract: Bacterial pathogens can evolve and diversify within hosts, leading to persistent infections that are highly challenging to treat. These evolutionary transitions can be difficult to detect even with state-of-the-art omics techniques, as multiple genetic changes can lead to the same phenotypic outcome. The opportunistic bacterial pathogen Pseudomonas aeruginosa, for example, causes persistent respiratory infections in patients with cystic fibrosis (CF). These infections cause progressive lung disease that severely limits their life-expectancy, with the median age at death estimated to be 30.8 years. Infections are usually established in early childhood, from environmental P. aeruginosa strains which do not pose huge risks to healthy individuals. Because they cannot be cleared by the immune system of CF sufferers, they persist in the lungs for many years. Over time they adapt to the environment of the lung, displaying new behaviours, or ‘pathoadaptations,’ which affect the severity of the disease. Infections become very difficult to treat, as bacteria become resistant to antibiotics and form extended biofilms.
I will describe an array of diagnostic molecular probes that can discriminate acute and chronic genotypes of P. aeruginosa based on phenotypic variation in their surface properties linked to important pathoadaptations. Using a combination of fluorescently labelled glycopolymers, this sensing array can distinguish genetically-engineered mutant strains differing in the expression of key virulence factors. The same sensing array can also be used to differentiate genetically variable P. aeruginosa isolates from CF patients sampled from different evolutionary lineages, and samples of the same lineage isolated from the same patient at different time points. This approach could provide the underpinning technology for new diagnostic tools to map the progress of persistent bacterial infections and inform treatment strategies.
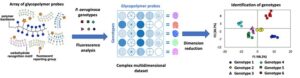
Graphical summary: Sensing arrays consisting of fluorescently-labelled glycopolymer probes can be used to distinguish P. aeruginosa genotypes based on phenotypic variation in their surface properties.

